LBF20207PG25: Difference between revisions
No edit summary |
No edit summary |
||
| Line 14: | Line 14: | ||
|IR Spectra=NEAT : 3320, 2640, 1710, 1295, 1260, 1245, 1120, 1080, 1055, 1025, 975cm<SUP><FONT SIZE=-1>-</FONT></SUP><SUP><FONT SIZE=-1>1</FONT></SUP> [[Reference:Pike_J_E:Lincoln_FH:Schneider_WP:,J. Org. Chem.,1969,34,3552|{{RelationTable/GetFirstAuthor|Reference:Pike_J_E:Lincoln_FH:Schneider_WP:,J. Org. Chem.,1969,34,3552}}]] | |IR Spectra=NEAT : 3320, 2640, 1710, 1295, 1260, 1245, 1120, 1080, 1055, 1025, 975cm<SUP><FONT SIZE=-1>-</FONT></SUP><SUP><FONT SIZE=-1>1</FONT></SUP> [[Reference:Pike_J_E:Lincoln_FH:Schneider_WP:,J. Org. Chem.,1969,34,3552|{{RelationTable/GetFirstAuthor|Reference:Pike_J_E:Lincoln_FH:Schneider_WP:,J. Org. Chem.,1969,34,3552}}]] | ||
|NMR Spectra=<SUP><FONT SIZE=-1>1</FONT></SUP>H-NMR(d<SUB><FONT SIZE=-1>6</FONT></SUB>-ACETONE) : <FONT FACE="Symbol">d</FONT> 5.48(m, 4H), 4.05(m, 3H), 0.9(t, 3H, 20-CH3) [[Reference:Pike_J_E:Lincoln_FH:Schneider_WP:,J. Org. Chem.,1969,34,3552|{{RelationTable/GetFirstAuthor|Reference:Pike_J_E:Lincoln_FH:Schneider_WP:,J. Org. Chem.,1969,34,3552}}]]. <SUP><FONT SIZE=-1>1</FONT></SUP><SUP><FONT SIZE=-1>3</FONT></SUP>C-NMR : 176.6(C1), 135.0(C14), 132.8(C5), 129.1(C13 or C6), 128.9(C6 or C13), 77.2(C11), 72.9(C15), 71.8(C9), 55.0(C12), 49.9(C8), 42,6(C10), 36.8(C16), 33.2(C2), 31,5(C18), 26.3(C4), 25.1(C7), 25.1(C17), 24.5 [[Reference:Lukacs_G:Piriou_F:Gero_SD:,Tetrah. Lett.,1973,,515|{{RelationTable/GetFirstAuthor|Reference:Lukacs_G:Piriou_F:Gero_SD:,Tetrah. Lett.,1973,,515}}]] | |NMR Spectra=<SUP><FONT SIZE=-1>1</FONT></SUP>H-NMR(d<SUB><FONT SIZE=-1>6</FONT></SUB>-ACETONE) : <FONT FACE="Symbol">d</FONT> 5.48(m, 4H), 4.05(m, 3H), 0.9(t, 3H, 20-CH3) [[Reference:Pike_J_E:Lincoln_FH:Schneider_WP:,J. Org. Chem.,1969,34,3552|{{RelationTable/GetFirstAuthor|Reference:Pike_J_E:Lincoln_FH:Schneider_WP:,J. Org. Chem.,1969,34,3552}}]]. <SUP><FONT SIZE=-1>1</FONT></SUP><SUP><FONT SIZE=-1>3</FONT></SUP>C-NMR : 176.6(C1), 135.0(C14), 132.8(C5), 129.1(C13 or C6), 128.9(C6 or C13), 77.2(C11), 72.9(C15), 71.8(C9), 55.0(C12), 49.9(C8), 42,6(C10), 36.8(C16), 33.2(C2), 31,5(C18), 26.3(C4), 25.1(C7), 25.1(C17), 24.5 [[Reference:Lukacs_G:Piriou_F:Gero_SD:,Tetrah. Lett.,1973,,515|{{RelationTable/GetFirstAuthor|Reference:Lukacs_G:Piriou_F:Gero_SD:,Tetrah. Lett.,1973,,515}}]] | ||
|Source=Prostaglandin F2<FONT FACE="Symbol">a</FONT> was found to be accummulating in human semen in an amount of about 2 microgram per ml [[Reference:Bergstrom_S:,Science,1967,157,382|{{RelationTable/GetFirstAuthor|Reference:Bergstrom_S:,Science,1967,157,382}}]] | |Source=Prostaglandin F2<FONT FACE="Symbol">a</FONT> was found to be accummulating in human semen in an amount of about 2 microgram per ml [[Reference:Bergstrom_S:,Science,1967,157,382|{{RelationTable/GetFirstAuthor|Reference:Bergstrom_S:,Science,1967,157,382}}]]. In most animal tissues prostanoids are synthesized enzymatically de novo upon physiological and pathological stimulations, and this is also the case of prostaglandin F2<FONT FACE="Symbol">a</FONT>. | ||
|Chemical Synthesis=[[Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397|{{RelationTable/GetFirstAuthor|Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397}}]] | |Chemical Synthesis=[[Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397|{{RelationTable/GetFirstAuthor|Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397}}]] {{Image200|LBF20207PG25FT0001.gif}} | ||
|Metabolism=Prostaglandin F synthase reduces 9,11-endoperoxide of prostaglandin H2 requiring NADPH, and produces prostaglandin F2<FONT FACE="Symbol">a</FONT>. The same enzyme also reduces 9-keto group of prostaglandin D2 producing 11<FONT FACE="Symbol">b</FONT>-prostaglandin F2 [[Reference:Urade_Y:Watanabe_K:Hayaishi_O:,J. Lipid Mediat. Cell Signal.,1995,12,257|{{RelationTable/GetFirstAuthor|Reference:Urade_Y:Watanabe_K:Hayaishi_O:,J. Lipid Mediat. Cell Signal.,1995,12,257}}]] | |Metabolism=Prostaglandin F synthase reduces 9,11-endoperoxide of prostaglandin H2 requiring NADPH, and produces prostaglandin F2<FONT FACE="Symbol">a</FONT>. The same enzyme also reduces 9-keto group of prostaglandin D2 producing 11<FONT FACE="Symbol">b</FONT>-prostaglandin F2 [[Reference:Urade_Y:Watanabe_K:Hayaishi_O:,J. Lipid Mediat. Cell Signal.,1995,12,257|{{RelationTable/GetFirstAuthor|Reference:Urade_Y:Watanabe_K:Hayaishi_O:,J. Lipid Mediat. Cell Signal.,1995,12,257}}]]. | ||
}} | }} | ||
{{Lipid/Footer}} | {{Lipid/Footer}} | ||
Revision as of 11:55, 25 November 2009
| LipidBank Top (トップ) |
Fatty acid (脂肪酸) |
Glycerolipid (グリセロ脂質) |
Sphingolipid (スフィンゴ脂質) |
Journals (雑誌一覧) |
How to edit (ページの書き方) |
| IDs and Links | |
|---|---|
| LipidBank | XPR1501 |
| LipidMaps | LMFA03010002 |
| CAS | |
| KEGG | {{{KEGG}}} |
| KNApSAcK | {{{KNApSAcK}}} |
| mol | LBF20207PG25 |
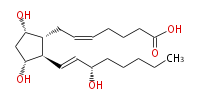
| PROSTAGLANDIN F_2α | |
|---|---|
| |
| Structural Information | |
| 7- [ 3 (R) ,5 (S) -Dihydroxy-2 (R) - (3 (S) -hydroxy-1 (E) -octenylcyclopentan-1 (R) -yl ] -5 (Z) -heptenoic acid | |
| |
| Formula | C20H34O5 |
| Exact Mass | 354.240624198 |
| Average Mass | 354.48096000000004 |
| SMILES | C(CC[C@@H](O)C=C[C@H]([C@H]1CC=CCCCC(O)=O)[C@@H](C[C@@H]1O)O)CC |
| Physicochemical Information | |
| 25-35°C Bundy_GL et al. | |
| ETHYL ACETATE, ACETONE, DIETHYLETHER Pike_JEet al.. STABILITIES: to be stable under neutral and basic conditions Karim_SM et al. | |
| Prostaglandin F2a was found to be accummulating in human semen in an amount of about 2 microgram per ml Bergstrom_S . In most animal tissues prostanoids are synthesized enzymatically de novo upon physiological and pathological stimulations, and this is also the case of prostaglandin F2a. | |
|
Corey_EJ et al. | |
| Prostaglandin F synthase reduces 9,11-endoperoxide of prostaglandin H2 requiring NADPH, and produces prostaglandin F2a. The same enzyme also reduces 9-keto group of prostaglandin D2 producing 11b-prostaglandin F2 Urade_Y et al.. | |
| Spectral Information | |
| Mass Spectra | 354(M+), 336, 318, 292, 274, 264(100), 247, 229, 191, 177, 165, 137, 99, 81, 67 HorvathG |
| UV Spectra | |
| IR Spectra | NEAT : 3320, 2640, 1710, 1295, 1260, 1245, 1120, 1080, 1055, 1025, 975cm-1 Pike_JEet al. |
| NMR Spectra | 1H-NMR(d6-ACETONE) : d 5.48(m, 4H), 4.05(m, 3H), 0.9(t, 3H, 20-CH3) Pike_JEet al.. 13C-NMR : 176.6(C1), 135.0(C14), 132.8(C5), 129.1(C13 or C6), 128.9(C6 or C13), 77.2(C11), 72.9(C15), 71.8(C9), 55.0(C12), 49.9(C8), 42,6(C10), 36.8(C16), 33.2(C2), 31,5(C18), 26.3(C4), 25.1(C7), 25.1(C17), 24.5 LukacsGet al. |
| Other Spectra | |
| Chromatograms | |
| Reported Metabolites, References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|