LBF20306HX04
| LipidBank Top (トップ) |
Fatty acid (脂肪酸) |
Glycerolipid (グリセロ脂質) |
Sphingolipid (スフィンゴ脂質) |
Journals (雑誌一覧) |
How to edit (ページの書き方) |
| IDs and Links | |
|---|---|
| LipidBank | XPR5101 |
| LipidMaps | LMFA03090003 |
| CAS | |
| KEGG | {{{KEGG}}} |
| KNApSAcK | {{{KNApSAcK}}} |
| mol | LBF20306HX04 |
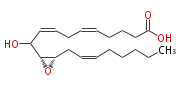
| Hepoxilin B3 | |
|---|---|
| |
| Structural Information | |
| 10-Hydroxy- (11R,12S)-epoxy- (cis-5,cis-8,cis-14) -eicosatrienoic acid | |
| |
| HxB3 | |
| Formula | C20H32O4 |
| Exact Mass | 336.23005951199997 |
| Average Mass | 336.46567999999996 |
| SMILES | C(CC=CC[C@@H]([C@@H](C(O)C=CCC=CCCCC(O)=O)1)O1)CCC |
| Physicochemical Information | |
| ACETATE, METHYL ESTER ; [ α ]X23 D =-10.9°(C=0.11, CHLOROFORM) Moghaddam_MF et al. | |
| DIETHYL ETHER Pace-Asciak_CR et al. | |
| Hepoxilin B3 together with hepoxilin A3 is produced from arachidonic acid or more directly from 12(S)-hydroperoxy-5,8,10,14-eicosatetraenoic acid in various animal tissues including brain, pineal gland, pancreas and skin Pace-Asciak_CR . | |
|
Corey_EJ et al. | |
| The presence of hepoxilin synthase was suggested by a finding that intact cells (skin) and tissue slices (brain hippocampus and pineal gland) transformed 12(S)-hydroperoxy-5,8,10,14-eicosatetraenoic acid to hepoxilins A3 and B3, but tissue boiling inhibited the hepoxilin production Pace-Asciak_CR et al.. | |
| Little has been described about the biological actvity of hepoxilin B3 except for its poteniation of glucose-dependent insulin secretion Pace-Asciak_CR and of bradykinin-induced increase of vascular permeability . | |
| Spectral Information | |
| Mass Spectra | METHYL ESTER TMS ETHER ; m/e 311, 282, 269(base peak) Moghaddam_MF et al. |
| UV Spectra | |
| IR Spectra | ACETATE METHYL ESTER ; ν (CHLOROFORM) 2956, 1743, 1550, 1372, 1234, 1033, 999cm-1 Moghaddam_MF et al. |
| NMR Spectra | ACETATE METHYL ESTER ; 1H-NMR(C6D6) : δ 5.67(dd, J=9.2, 6.4Hz, 1H, 10-CH), 5.52, 5.46, 5.42, 5.35, 5.32, 3.36(s, 3H, OCH3), 2.95(m, 2H, 7-CH), 2.92(m, 1H, 11-CH), 2.86(ddd, J=7.4, 7.4, 2.1Hz, 1H, 12-CH), 2.30(ddd, J=14.8, 7.4, 7.4Hz, 1H, 13-CH), 2.18(ddd, J=14.8, 7.4, 7.4Hz, 1H, 13-CH), 2.12(t, J=7.4Hz, 2H, 2-CH), 1.98(dt, J=7.4, 7.4Hz, 2H, 4-CH), 1.92(dt, J=8.8, 8.8Hz, 2H, 16-CH), 1.65(s, 3H, COCH3), 1.60(tt, J=7.4, 7.4Hz, 2H, 3-CH), 1.25(m, 6H), 0.88(t, J=7.0Hz, 3H, 20-CH). Moghaddam_MF et al. 13NMR(C6D6) : 134.22, 133.35, 130.17, 127.74, 124.31, 123.37, 70.86, 58.30, 55.71, 50.94, 33.30, 31.71, 29.66, 29.52, 27.59, 26.73, 25.00, 22.88, 20.53, 14.23 Moghaddam_MF et al. |
| Other Spectra | |
| Chromatograms | |
| Reported Metabolites, References | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|