LBF20406LT02
| LipidBank Top (トップ) |
Fatty acid (脂肪酸) |
Glycerolipid (グリセロ脂質) |
Sphingolipid (スフィンゴ脂質) |
Journals (雑誌一覧) |
How to edit (ページの書き方) |
| IDs and Links | |
|---|---|
| LipidBank | XPR3101 |
| LipidMaps | LMFA03020001 |
| CAS | |
| KEGG | {{{KEGG}}} |
| KNApSAcK | {{{KNApSAcK}}} |
| mol | LBF20406LT02 |
| LEUKOTRIENE B4 | |
|---|---|
| |
| Structural Information | |
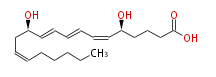
| 5 (S) ,12 (R) -Dihydroxyeicosa-6 (Z) ,8 (E) ,10 (E) ,14 (Z) -tetraenoic acid | |
| |
| LTB4 | |
| Formula | C20H32O4 |
| Exact Mass | 336.23005951199997 |
| Average Mass | 336.46567999999996 |
| SMILES | C(CC=CC[C@H](C=CC=CC=C[C@H](CCCC(O)=O)O)O)CCC |
| Physicochemical Information | |
| METHANOL Corey_EJ et al. | |
| Leukotriene B4 is produced by polymorphonuclear leukocytes and macrophages of various animal species upon various stimulations on the cells Samuelsson_B et al. Hammarstrom_S . | |
Corey_EJ et al. 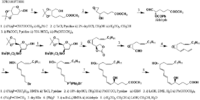 | |
| Arachidonic acid is metabolized to leukotrienen A4 with 5,6-epoxide by 5-lipoxygenases, and the product is further transformed to leukotrienee B4 by leukotriene A hydrolase Ford-Hutchinson_AW et al.. Leukotriene B4 is metabolized to lose its bioactivities either by omega -oxidation Hammarstrom_S or by leukotriene B4 12-hydroxydehydrogenase Yokomizo_T et al.. | |
| Leukotriene B4 causes adhesion of leukocytes to endothelial cells, stimulates chemotaxis and chemokinesis of leukocytes Hammarstrom_S , and enhances superoxide anion production by human polymorphonuclear leukocytes Sumimoto_H et al.. Leukotriene B4 binds to a specific receptor with 7 transmembrane domains coupled to Gi/Go or Gq protein Yokomizo_T et al.. | |
| cDNA and genomic DNA of 5-lipoxygenase Funk_CD and cDNA for Leukotriene A hydrolase Funk_CD were cloned. cDNA for leukotriene B 12-hydroxydehydrogenase was cloned Yokomizo_T et al.. | |
| Spectral Information | |
| Mass Spectra | m/e 336, 319, 301 Yergey_JA et al. |
| UV Spectra | METHANOL : 260( ε 38,000), 270.5( ε 50,000), 281( ε 39,000)nm Corey_EJ et al. |
| IR Spectra | |
| NMR Spectra | 1H-NMR(250MHz, D2O) : δ 6.45(m, 1H, 8-CH), 6.15(m, 2H, 9,10-CH), 6.0(m, 1H, 7-CH), 5.65(m, 1H, 11-CH), 5.40(m, 1H, 15-CH), 5.25(m, 2H, 6,14-CH), 4.60(5-CH), 4.05(m, 1H, 12-CH), 2.15(m, 2H, 13-CH), 2.00(m, 1H, 2-CH), 1.85(m, 2H, 16-CH), 1.35-1.60(m, 4H, 3,4-CH), 1.00-1.25(m, 6H, 17,18,19-CH), 0.70(m, 3H, 20-CH) Merrer_YL et al. |
| Other Spectra | |
| Chromatograms | |
| Reported Metabolites, References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|