LBF20207PG23: Difference between revisions
No edit summary |
No edit summary |
||
| (11 intermediate revisions by 2 users not shown) | |||
| Line 6: | Line 6: | ||
|LipidBank=XPR1401 | |LipidBank=XPR1401 | ||
|LipidMaps=LMFA03010003 | |LipidMaps=LMFA03010003 | ||
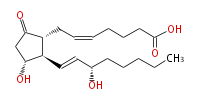
|SysName=7- [ | |SysName=7- [3R -Hydroxy-2R - (3S -hydroxy-trans-1-octenyl-5-oxocyclopentan-1R -yl] -cis-5-heptenoic acid | ||
|Common Name=&&Prostaglandin E_2&&7- [ 3 (R) -Hydroxy-2 (R) - (3 (S) -hydroxy-1 (E) -octenyl-5-oxocyclopentan-1 (R) -yl ] -5 (Z) -heptenoic acid | |Common Name=&&Prostaglandin E_2&&7- [ 3 (R) -Hydroxy-2 (R) - (3 (S) -hydroxy-1 (E) -octenyl-5-oxocyclopentan-1 (R) -yl ] -5 (Z) -heptenoic acid | ||
|Melting Point=65-66°C [[Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397|{{RelationTable/GetFirstAuthor|Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397}}]] | |Melting Point=65-66°C [[Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397|{{RelationTable/GetFirstAuthor|Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397}}]] | ||
| | |Optical=[ alpha ]^{26}_D =-61.0°(C=1.0, TETRAHYDROFURAN) [[Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397|{{RelationTable/GetFirstAuthor|Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397}}]] | ||
|Solubility= ETHYL ACETATE,THF,CHLOROFORM[[Reference:Donaldson_RE:Saddler_JC:Byrn_S:McKenzie_AT:Fuchs_PL:,J. Org. Chem.,1983,48,2167|{{RelationTable/GetFirstAuthor|Reference:Donaldson_RE:Saddler_JC:Byrn_S:McKenzie_AT:Fuchs_PL:,J. Org. Chem.,1983,48,2167}}]][[Reference:Sih_CJ:Heather_JB:Sood_R:Price_P:Peruzzotti_G:Lee_LF:Lee_SS:,J. Am. Chem. Soc.,1975,97,865|{{RelationTable/GetFirstAuthor|Reference:Sih_CJ:Heather_JB:Sood_R:Price_P:Peruzzotti_G:Lee_LF:Lee_SS:,J. Am. Chem. Soc.,1975,97,865}}]]. STABILITIES: to be stable under neutral condition. to decompose to PGA2 under acidic and to PGB2 under basic conditions [[Reference:Karim_SM:Devlin_J:Hillier_K:,Eur. J. Pharmacol.,1968,4,416|{{RelationTable/GetFirstAuthor|Reference:Karim_SM:Devlin_J:Hillier_K:,Eur. J. Pharmacol.,1968,4,416}}]][[Reference: | |Solubility= ETHYL ACETATE,THF,CHLOROFORM[[Reference:Donaldson_RE:Saddler_JC:Byrn_S:McKenzie_AT:Fuchs_PL:,J. Org. Chem.,1983,48,2167|{{RelationTable/GetFirstAuthor|Reference:Donaldson_RE:Saddler_JC:Byrn_S:McKenzie_AT:Fuchs_PL:,J. Org. Chem.,1983,48,2167}}]][[Reference:Sih_CJ:Heather_JB:Sood_R:Price_P:Peruzzotti_G:Lee_LF:Lee_SS:,J. Am. Chem. Soc.,1975,97,865|{{RelationTable/GetFirstAuthor|Reference:Sih_CJ:Heather_JB:Sood_R:Price_P:Peruzzotti_G:Lee_LF:Lee_SS:,J. Am. Chem. Soc.,1975,97,865}}]]. STABILITIES: to be stable under neutral condition. to decompose to PGA2 under acidic and to PGB2 under basic conditions [[Reference:Karim_SM:Devlin_J:Hillier_K:,Eur. J. Pharmacol.,1968,4,416|{{RelationTable/GetFirstAuthor|Reference:Karim_SM:Devlin_J:Hillier_K:,Eur. J. Pharmacol.,1968,4,416}}]][[Reference:Pike_JE:Lincoln_FH:Schneider_WP:,J. Org. Chem.,1969,34,3552|{{RelationTable/GetFirstAuthor|Reference:Pike_JE:Lincoln_FH:Schneider_WP:,J. Org. Chem.,1969,34,3552}}]] | ||
|Mass Spectra=d,l-mixture ; 334(M | |Mass Spectra=d,l-mixture ; 334(M^+ -18), 316, 298, 190 [[Reference:Chen_SML:Schaub_RE:Grudzinskas_CV:,J. Org. Chem.,1978,43,3450|{{RelationTable/GetFirstAuthor|Reference:Chen_SML:Schaub_RE:Grudzinskas_CV:,J. Org. Chem.,1978,43,3450}}]] | ||
|IR Spectra=d,l-mixture ; 3400, 1710, 970cm | |IR Spectra=d,l-mixture ; 3400, 1710, 970cm^{-1} [[Reference:Chen_SML:Schaub_RE:Grudzinskas_CV:,J. Org. Chem.,1978,43,3450|{{RelationTable/GetFirstAuthor|Reference:Chen_SML:Schaub_RE:Grudzinskas_CV:,J. Org. Chem.,1978,43,3450}}]] | ||
|NMR Spectra= | |NMR Spectra=^{13}C-NMR(CDCl_3 ) : 214.71(C9),178.39(C1), 136.62(C14), 131.52(C13), 130.91(C5), 126.69(C6), 73.19(C15), 72.13(C11), 54.55(C12), 53.51(C8), 46.23(C10), 37.00 (C16), 33.56(C2), 31.73(C18), 26.47(C4), 25.20(C7,17), 24.60(C3), 22.64(C19), 14.04. [[Reference:Donaldson_RE:Saddler_JC:Byrn_S:McKenzie_AT:Fuchs_PL:,J. Org. Chem.,1983,48,2167|{{RelationTable/GetFirstAuthor|Reference:Donaldson_RE:Saddler_JC:Byrn_S:McKenzie_AT:Fuchs_PL:,J. Org. Chem.,1983,48,2167}}]]. ^1 H-NMR(CDCl_3 ) : delta 5.67(dd, J=6.6Hz, 15.4, 1H, 14-CH), 5.57(dd, J=8.1, 15.4Hz, 1H, 13-CH), 5.40(m, 2H, 5.6-CH), 4.12(q, J=6.5, 6.7, 6.8Hz, 1H, 15-CH), 4.06(q, J=8.1, 8.2, 8.3Hz, 1H, 11-CH), 2.72(dd, J= [[Reference:Donaldson_RE:Saddler_JC:Byrn_S:McKenzie_AT:Fuchs_PL:,J. Org. Chem.,1983,48,2167|{{RelationTable/GetFirstAuthor|Reference:Donaldson_RE:Saddler_JC:Byrn_S:McKenzie_AT:Fuchs_PL:,J. Org. Chem.,1983,48,2167}}]] | ||
|Source=Prostaglandin E2 was found to be accummulating in human semen in an amount of about 13 microgram per ml [[Reference:Bergstrom_S:,Science,1967,157,382|{{RelationTable/GetFirstAuthor|Reference:Bergstrom_S:,Science,1967,157,382}}]]. In most animal tissues prostanoids are synthesized enzymatically de novo upon physiological and pathological stimulations, and this is also the case of prostaglandin E2. | |Source=Prostaglandin E2 was found to be accummulating in human semen in an amount of about 13 microgram per ml [[Reference:Bergstrom_S:,Science,1967,157,382|{{RelationTable/GetFirstAuthor|Reference:Bergstrom_S:,Science,1967,157,382}}]]. In most animal tissues prostanoids are synthesized enzymatically de novo upon physiological and pathological stimulations, and this is also the case of prostaglandin E2. | ||
|Chemical Synthesis={{Image200|LBF20207PG23FT0001.gif}} [[Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397|{{RelationTable/GetFirstAuthor|Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397}}]] | |Chemical Synthesis={{Image200|LBF20207PG23FT0001.gif}} [[Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397|{{RelationTable/GetFirstAuthor|Reference:Corey_EJ:Schaaf_TK:Huber_W:Koelliker_U:Weinshenker_NM:,J. Am. Chem. Soc.,1970,92,397}}]] | ||
|Metabolism=Prostaglandin E2 is produced from arachidonic acid via prostaglandins G2 and H2 by the catalyses of prostaglandin endoperoxide synthase (cyclooxygenase) [[Reference:Smith_WL:Garavito_RM:DeWitt_DL:,J. Biol. Chem.,1996,271,33157|{{RelationTable/GetFirstAuthor|Reference:Smith_WL:Garavito_RM:DeWitt_DL:,J. Biol. Chem.,1996,271,33157}}]] and prostaglandin E synthase [[Reference:Urade_Y:Watanabe_K:Hayaishi_O:,J. Lipid Mediat. Cell Signal.,1995,12,257|{{RelationTable/GetFirstAuthor|Reference:Urade_Y:Watanabe_K:Hayaishi_O:,J. Lipid Mediat. Cell Signal.,1995,12,257}}]]. Two isoforms of the cyclooxygnease enzyme responsible for prostaglandin H2 synthesis are present. Cyclooxygenase-1 is constitutively found in most mammalian tissues, while cyclooxygnease-2 is induced rapidly and transiently in physiological and pathological events, especially in inflammation [[Reference:Smith_WL:Garavito_RM:DeWitt_DL:,J. Biol. Chem.,1996,271,33157|{{RelationTable/GetFirstAuthor|Reference:Smith_WL:Garavito_RM:DeWitt_DL:,J. Biol. Chem.,1996,271,33157}}]]. The biological activities of prostaglandin E2 are lost by oxidation of its 15-hydroxyl group catalyzed by 15-hydroxyprostaglandin dehydrogenase [[Reference:Hansen_HS:,Prostaglandins,1976,12,647|{{RelationTable/GetFirstAuthor|Reference:Hansen_HS:,Prostaglandins,1976,12,647}}]]. | |Metabolism=Prostaglandin E2 is produced from arachidonic acid via prostaglandins G2 and H2 by the catalyses of prostaglandin endoperoxide synthase (cyclooxygenase) [[Reference:Smith_WL:Garavito_RM:DeWitt_DL:,J. Biol. Chem.,1996,271,33157|{{RelationTable/GetFirstAuthor|Reference:Smith_WL:Garavito_RM:DeWitt_DL:,J. Biol. Chem.,1996,271,33157}}]] and prostaglandin E synthase [[Reference:Urade_Y:Watanabe_K:Hayaishi_O:,J. Lipid Mediat. Cell Signal.,1995,12,257|{{RelationTable/GetFirstAuthor|Reference:Urade_Y:Watanabe_K:Hayaishi_O:,J. Lipid Mediat. Cell Signal.,1995,12,257}}]]. Two isoforms of the cyclooxygnease enzyme responsible for prostaglandin H2 synthesis are present. Cyclooxygenase-1 is constitutively found in most mammalian tissues, while cyclooxygnease-2 is induced rapidly and transiently in physiological and pathological events, especially in inflammation [[Reference:Smith_WL:Garavito_RM:DeWitt_DL:,J. Biol. Chem.,1996,271,33157|{{RelationTable/GetFirstAuthor|Reference:Smith_WL:Garavito_RM:DeWitt_DL:,J. Biol. Chem.,1996,271,33157}}]]. The biological activities of prostaglandin E2 are lost by oxidation of its 15-hydroxyl group catalyzed by 15-hydroxyprostaglandin dehydrogenase [[Reference:Hansen_HS:,Prostaglandins,1976,12,647|{{RelationTable/GetFirstAuthor|Reference:Hansen_HS:,Prostaglandins,1976,12,647}}]]. | ||
|Symbol=PGE2 | |||
|Biological Activity=Prostaglandin E2 exhibits various biological activities such as vasodilatation, uterine contraction, gastrointestinal contraction, bronchodilatation, diuresis, pyrexia, inhibition of gastric secretion, bone resorption and immunosuppression [[Reference:Bergstrom_S:Carlson_LA:Weeks_JR:,Pharmacol. Rev.,1968,20,1|{{RelationTable/GetFirstAuthor|Reference:Bergstrom_S:Carlson_LA:Weeks_JR:,Pharmacol. Rev.,1968,20,1}}]]. Prostaglandin E2 is a ligand to receptors present in the cell membrane, and there are at least 4 subtypes of its receptor. Different tissue distributions and signal transductions of these receptor subtypes explain a variety of biological activities of prostaglandin E2 [[Reference:Negishi_M:Sugimoto_Y:Ichikawa_A:,Biochim. Biophys. Acta,1995,1259,109|{{RelationTable/GetFirstAuthor|Reference:Negishi_M:Sugimoto_Y:Ichikawa_A:,Biochim. Biophys. Acta,1995,1259,109}}]]. | |||
|Genetic Information=cDNAs of the two cyclooxygenase isozymes responsible for prostaglandin H2 synthesis have been cloned, and the primary structures of these enzymes were deduced from the nucleotide seuences [[Reference:Funk_CD:,Prog Nucleic Acid Res Mol Biol.,1993,45,67|{{RelationTable/GetFirstAuthor|Reference:Funk_CD:,Prog Nucleic Acid Res Mol Biol.,1993,45,67}}]][[Reference:Herschman_HR:,Biochim._Biophys._Acta,1996,1299,125|{{RelationTable/GetFirstAuthor|Reference:Herschman_HR:,Biochim._Biophys._Acta,1996,1299,125}}]] Their genomic DNA were also cloned, and the genomic structure were eludicated [[Reference:Funk_CD:,Prog Nucleic Acid Res Mol Biol.,1993,45,67|{{RelationTable/GetFirstAuthor|Reference:Funk_CD:,Prog Nucleic Acid Res Mol Biol.,1993,45,67}}]]. cDNAs of four subtypes of prostaglandin E2 receptors (EP1-4) were cloned, and the 7-transmembrane domain structures of the receptors coupled with certain G proteins were reported [[Reference:Negishi_M:Sugimoto_Y:Ichikawa_A:,Biochim. Biophys. Acta,1995,1259,109|{{RelationTable/GetFirstAuthor|Reference:Negishi_M:Sugimoto_Y:Ichikawa_A:,Biochim. Biophys. Acta,1995,1259,109}}]]. | |||
}} | |||
{{MassbankSpectra| | |||
UT000343 | |||
UT000344 | |||
UT000345 | |||
UT000346 | |||
UT000347 | |||
UT000348 | |||
UT000349 | |||
UT000350 | |||
UT000351 | |||
}} | }} | ||
{{Lipid/Footer}} | {{Lipid/Footer}} | ||
Latest revision as of 09:00, 17 January 2014
| LipidBank Top (トップ) |
Fatty acid (脂肪酸) |
Glycerolipid (グリセロ脂質) |
Sphingolipid (スフィンゴ脂質) |
Journals (雑誌一覧) |
How to edit (ページの書き方) |
| IDs and Links | |
|---|---|
| LipidBank | XPR1401 |
| LipidMaps | LMFA03010003 |
| CAS | |
| KEGG | {{{KEGG}}} |
| KNApSAcK | {{{KNApSAcK}}} |
| mol | LBF20207PG23 |
| Prostaglandin E2 | |
|---|---|
| |
| Structural Information | |
| 7- [3R -Hydroxy-2R - (3S -hydroxy-trans-1-octenyl-5-oxocyclopentan-1R -yl] -cis-5-heptenoic acid | |
| |
| PGE2 | |
| Formula | C20H32O5 |
| Exact Mass | 352.224974134 |
| Average Mass | 352.46508 |
| SMILES | C(CC[C@@H](O)C=C[C@H]([C@H]1CC=CCCCC(O)=O)[C@@H](CC1=O)O)CC |
| Physicochemical Information | |
| 65-66°C Corey_EJ et al. | |
| [ α ]26 D =-61.0°(C=1.0, TETRAHYDROFURAN) Corey_EJ et al. | |
| ETHYL ACETATE,THF,CHLOROFORM Donaldson_RE et al. Sih_CJ et al.. STABILITIES: to be stable under neutral condition. to decompose to PGA2 under acidic and to PGB2 under basic conditions Karim_SM et al. Pike_JE et al. | |
| Prostaglandin E2 was found to be accummulating in human semen in an amount of about 13 microgram per ml Bergstrom_S . In most animal tissues prostanoids are synthesized enzymatically de novo upon physiological and pathological stimulations, and this is also the case of prostaglandin E2. | |
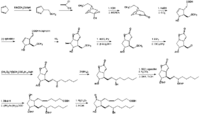 Corey_EJ et al. | |
| Prostaglandin E2 is produced from arachidonic acid via prostaglandins G2 and H2 by the catalyses of prostaglandin endoperoxide synthase (cyclooxygenase) Smith_WL et al. and prostaglandin E synthase Urade_Y et al.. Two isoforms of the cyclooxygnease enzyme responsible for prostaglandin H2 synthesis are present. Cyclooxygenase-1 is constitutively found in most mammalian tissues, while cyclooxygnease-2 is induced rapidly and transiently in physiological and pathological events, especially in inflammation Smith_WL et al.. The biological activities of prostaglandin E2 are lost by oxidation of its 15-hydroxyl group catalyzed by 15-hydroxyprostaglandin dehydrogenase Hansen_HS . | |
| Prostaglandin E2 exhibits various biological activities such as vasodilatation, uterine contraction, gastrointestinal contraction, bronchodilatation, diuresis, pyrexia, inhibition of gastric secretion, bone resorption and immunosuppression Bergstrom_S et al.. Prostaglandin E2 is a ligand to receptors present in the cell membrane, and there are at least 4 subtypes of its receptor. Different tissue distributions and signal transductions of these receptor subtypes explain a variety of biological activities of prostaglandin E2 Negishi_M et al.. | |
| cDNAs of the two cyclooxygenase isozymes responsible for prostaglandin H2 synthesis have been cloned, and the primary structures of these enzymes were deduced from the nucleotide seuences Funk_CD Herschman_HR Their genomic DNA were also cloned, and the genomic structure were eludicated Funk_CD . cDNAs of four subtypes of prostaglandin E2 receptors (EP1-4) were cloned, and the 7-transmembrane domain structures of the receptors coupled with certain G proteins were reported Negishi_M et al.. | |
| Spectral Information | |
| Mass Spectra | d,l-mixture ; 334(M+-18), 316, 298, 190 Chen_SML et al. |
| UV Spectra | |
| IR Spectra | d,l-mixture ; 3400, 1710, 970cm-1 Chen_SML et al. |
| NMR Spectra | 13C-NMR(CDCl3) : 214.71(C9),178.39(C1), 136.62(C14), 131.52(C13), 130.91(C5), 126.69(C6), 73.19(C15), 72.13(C11), 54.55(C12), 53.51(C8), 46.23(C10), 37.00 (C16), 33.56(C2), 31.73(C18), 26.47(C4), 25.20(C7,17), 24.60(C3), 22.64(C19), 14.04. Donaldson_RE et al.. 1H-NMR(CDCl3) : δ 5.67(dd, J=6.6Hz, 15.4, 1H, 14-CH), 5.57(dd, J=8.1, 15.4Hz, 1H, 13-CH), 5.40(m, 2H, 5.6-CH), 4.12(q, J=6.5, 6.7, 6.8Hz, 1H, 15-CH), 4.06(q, J=8.1, 8.2, 8.3Hz, 1H, 11-CH), 2.72(dd, J= Donaldson_RE et al. |
| Other Spectra | |
| Chromatograms | |
| Reported Metabolites, References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|








